
ERC Advanced Grant “Symbiotics” (2011-2017)
Objectives
- To discover the activities and modes of action of symbiotic plant peptides in differentiation of nitrogen fixing bacteria
- To reveal the significance of endoploidy in the plant and bacterial symbionts
- To determine and describe the antimicrobial activity (spectrum, MIC,) of symbiotic plant peptides on selected bacteria, fungi and to identify their intracellular targets
- To initiate biotech applications: Sym-Biotics drug development, testing the potential of Sym-Biotics for plant protection, food safety (food processing industry) and human/animal therapies.
Results
1. The symbiotic role of NCR peptides
In the indeterminate M. truncatula nodules there is an age gradient from the apical meristem to the nitrogen-fixing cells which represents all stages of symbiotic cell development (Kondorosi et al 2013, Maróti and Kondorosi, 2014, Ábrahám et al 2015). The small non-dividing cells leaving the meristem are infected with the bacteria, which are then gradually differentiate into nitrogen-fixing symbiotic cells along ~15 cell layers during their ageing. During this process the symbiotic nodule cells grow by repeated duplication of their genomes and the resulting polyploidy is essential for nodule differentiation. The endosymbionts multiply in the freshly infected nodule cells but after a given bacterial cell density and growth of the host cell, they permanently lose their ability to divide, and similarly to the host cell, they amplify their genome resulting in the formation of non-cultivable polyploid bacteria.
1.1 Extreme nodule specific expression and transcriptional waves of NCR genes correlate with ploidy dependent changes in the epigenome.
Expression of NCR genes is subject to an extremely tight regulation as they are only activated by the presence of endosymbionts during nodule organogenesis. This expression occurs in successive waves from the apical to the proximal nodule zones (Guefrachi et al. 2014). Moreover, their expression level is among the highest of all genes. We have shown that expression of early genes is coupled to lower while that of later genes to higher ploidy levels. While DNA methylation was unaffected by the ploidy levels and was independent of the active or repressed state of most genes, expression of nodule-specific genes correlated with ploidy-dependent opening of the chromatin and with reduced H3K27me3 levels, combined with enhanced H3K9ac levels (Nagymihály et al. 2017). This was the first study on ploidy dependent changes in the epigenome.
1.2 Why does the host cell produce an arsenal of NCRs? What can be the advantage of such a diverse peptide repertoire? Is it necessary for the interaction of the host with various bacteria?
The symbiotic partners of M. truncatula are S. meliloti and S. medicae, however in the soil there are countless strain variants of both species. M. truncatula is also represented by many different ecotypes and accessions differing in the number, sequences, and expression profiles of NCR genes (Nallu et al., 2014; Roux et al., 2014) and in their symbiotic interactions with different S. meliloti and S. medicae strains (Kazmierczak et al. 2017). In a collaborative work we have shown that an NCR peptide enforces symbiotic selectivity in M. truncatula (Wang et al, 2017). Thus NCR peptides in M. truncatula and possibly in closely related IRLC legumes could provide another level of host determinants of symbiotic specificity.
1.3 Diverse morphotypes of bacteroids in different IRLC legumes correlate with the number and type of NCR peptides
The fate of bacteroids in M. truncatula and other IRLC legumes is irreversible due to the loss of cell division capacity. Nevertheless, the morphology of the endosymbionts shows great variations, such as being swollen, elongated, spherical or elongated-branched (Montiel et al. 2016). Selecting 10 legumes, representing different subclades of the IRLC with distinct bacteroid morphotypes, we identified their NCR gene repertoire ranging from 7 up to >600. Analyzing the expression and predicted sequences of the NCRs established correlations between the complexity of the NCR families and the morphotypes of bacteroids (Montiel et al. 2017). Enrichment and diversification of cationic peptides had particular impact on the elongated-branched morphology of bacteroids.
1.4 Interaction of M. truncatula NCR peptides with bacteria
Interaction with the bacterial membranes
Mature NCR peptides show high diversity in their amino acid sequence and composition, consequently their isoelectric point varies from pI=3.2 to pI=11.2. Generally cationic peptides can interact with negatively charged bacterial membranes. Indeed, cationic NCR peptides (pI >9.2) but not the neutral or anionic peptides interact with the bacterial membranes. The outcome of the interactions is dose dependent. Using synthetic fluorochrome (FITC) labeled NCR peptides at low (sublethal) concentrations, certain cationic peptides were found in the bacterial membranes, while others in the cytosol (Fig. 1.). These latter self-penetrating peptides at this low concentration did not damage the membrane since propidium iodine (PI) could not enter the bacteria (Farkas et al. 2014). However, increasing the peptide concentrations (2.5-50 µM) provoked damage of the bacterial cell envelope (Nagy et al. 2015). The final outcome varied by the peptides’ physicochemical properties from surface roughness to complete cell disruption (Farkas et al. 2017). In many cases, budding of cells, formation of outer membrane vesicles or filaments were observed (Montiel et al. 2017). On model membranes strong lipid preference of NCR247 and NCR335 was observed suggesting that the bacterial cell envelope, especially the lipid matrix could be their first target (Nagy et al. 2015). NCR247 and NCR335 damaged both the outer and inner membranes but at different extent and resulted in the loss of membrane potential (Mikuláss et al, 2016). Treatment of S. meliloti cultures with either peptide, provoked quick downregulation of genes involved in basic cellular functions (transcription, translation, energy production) and up-regulation of genes involved in stress responses (Tiricz et al. 2013).

Figure 1.: Localization of NCR peptides (green fluorescence) in bacterial membranes (A) and in the cytosol of nitrogen fixing bacteroids.
However, in planta NCRs do not kill their bacterium partners. In the nodules, NCRs are likely produced at much lower concentrations and probably the combined action of anionic and neutral peptides can mitigate the antimicrobial properties of the cationic NCRs. Moreover, on the bacterium side the bacA gene proved to be essential for survival of endosymbionts exposed to NCRs (Guefrachi et al. 2015).
Numerous intracellular bacterial targets
The NCRs are delivered in subsequent waves to the endosymbionts. While cationic NCRs can pass through the bacterial membranes it is still unknown how non-cationic peptides enter the bacteria. Nevertheless, our large scale detection of NCRs from nodules and isolated endosymbionts revealed the presence of ~150 plant NCR peptides in the bacteroids with predominance of anionic and neutral peptides (Dürgo et al. 2015). Moreover, the presence of early NCRs in the mature nitrogen-fixing bacteroids indicated their high stability and possibly persisting biological roles in the bacteroids.
One of the shortest M. truncatula NCR peptides is NCR247. It consists of 24 amino acids, and based on its physicochemical properties, it can have extremely high protein-binding capacity. As this peptide accumulates in the bacterial cytosol, we looked for its bacterial targets (Farkas et al. 2014). The NCR247 gene is expressed when bacterial cell division is arrested and cell elongation begins. Its binding to FtsZ abolished septum formation and thus bacterial cell division and resulted in cell elongation. Complex formation with ribosomal proteins contributed to the altered proteome and physiology of the endosymbiont. Moreover, its binding to the chaperone GroEL amplified the NCR247-modulated biological processes. Thus NCR247 and likely many other NCRs have multiple bacterial targets and their combined action might affect specific biological pathways at several points.
Individual NCR peptides can be essential
Due to the expansion of the NCR family by gene duplications, probably many gene functions are redundant. However, our work showed that loss of the NCR169 gene in the M. truncatula dnf7 mutant abolished symbiotic nitrogen fixation, demonstrating that specific NCR peptides can have essential, non-redundant functions (Horváth et al. 2015).
2. Exploring the biological activities of the peptides beyond symbiosis, especially their antimicrobial potential for development of novel peptide-based antibiotics
2.1 Cationic NCR peptides: potent killers of various Gram positive and Gram negative bacteria and fungi
Antimicrobial peptides with broad antimicrobial activities represent one of the first lines of defense against pathogens. Many plant cysteine-rich peptides with possible antimicrobial properties have been predicted and amongst them, defensins and defensin-like peptides are the most abundant in plants. NCRs resemble to certain extent to defensins and as several cationic NCR peptides prevented growth of S. meliloti and killed them at higher concentrations, it was likely that cationic NCRs will effectively eliminate other microbes as well. Due to the alarming rise of infectious diseases, spread of antibiotic resistant bacteria, and lack of effective treatments, the Sym-Biotics project focused on the NCRs’ antimicrobial potential. 40 different synthetic NCR peptides were tested on various Gram positive and Gram negative bacteria and 16 fungal strains. NCR335 and NCR247 have the broadest antimicrobial activities; however, their bactericidal spectrum was not identical (Tiricz et al. 2013). While NCR247 completely eliminated within 3 hours 108 cells of S. typhimurium and S. aureus, NCR335 was nearly ineffective against these bacteria. In contrast, NCR335 effectively killed E. coli and P. aeruginosa what NCR247 did not, indicating that in addition to the positive charge, the amino acid composition and sequence also contribute to the antimicrobial activity. The effect of NCR335, NCR247, polymyxin B (PMB) and streptomycin on S. enterica and L. monocytogenes was also investigated and compared (Farkas et al. 2017, Fig. 2.). NCR335 and NCR247 accumulated in the cytosol of S. enterica and in the membranes of L. monocytogenes. NCR335 disrupted both bacteria while NCR247 provoked extensive budding on the cell surface of S. enterica. PMB had no effect on L. monocytogenes but effectively killed S. enterica while streptomycin was ineffective on both bacteria.
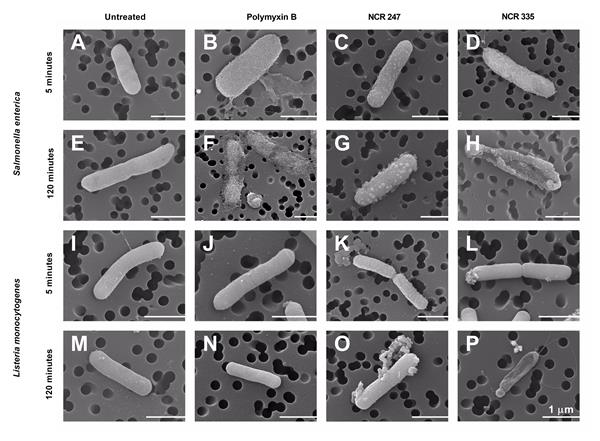
Figure 2.: Scanning electron microscopy of peptide-treated bacteria
Peptides with pI>9.5 exhibited anticandidal activity against growing Candida albicans, C. krusei, C. parapsilosis strains (Ördögh et al. 2014). The minimal inhibitory concentration of the peptides ranged from 1.42 µM to 10.5 µM that was comparable to that of the antifungal amphotericin B (1.69 µM). The peptides, particularly NCR247, NCR335 and NCR192 increased the membrane permeability of C. albicans cells and after 2 hrs of treatment caused drastic destruction of the cell membranes. Low concentrations of cationic NCRs efficiently eliminated C. albicans without affecting the viability of vaginal epithelial cells and thus the peptides inhibited C. albicans-induced killing of these cells (Ördögh et al. 2014). NCR247 and NCR335 showed antifungal activity on all tested fungal species and strains while other peptides had more restricted, specific activities (Ördögh L. PhD thesis).
2.2 NCRs are not toxic for human/animal cell lines The effects of NCRs on cell proliferation and cell adhesion as well as their cytotoxicity were studied on 7 human, 1 mouse and 1 insect cell lines. Treatment of the cell cultures with any of the tested peptides in culture media at 6 µM concentration provoked no cytotoxicity.