|
Principal Investigator: Pál Ormos
We developed a method that offers the possibility to directly apply and measure torque on
particles in an optical trap. It can be used to rotationally manipulate biopolymers attached to
appropriate particles. A flat object is trapped and oriented in the focus of a linearly polarized
laser light. The direction and power of the orientational trap are controlled by the polarization
state of the light. As a demonstration of the capabilities of the method, we examined the
torsional stiffness of dsDNA (l-DNA) in its linear torsional regime by directly measuring the
torque generated by the molecule.
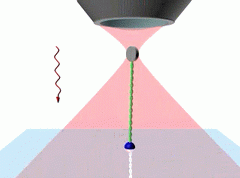
The Measurement In our system the molecule is attached to the trapped object (a disc shaped plastic particle, 2 µm) by one end and to a fixed plastic surface by the other end. The linearly polarized trapping light has an orienting effect on the flat test particle. By rotating the plane of polarization, the orientation of the disc can be rotated, thus the attached molecule can be twisted and torsionally stressed. As turns are added, the orientation of the disc drops behind the plane of polarization, since the torsional strain of the molecule acts against the orienting power of the light. By measuring the relevant angles (using video analysis) the molecular torsional stiffness can be directly compared to the orientational trapping power of the light. After calibrating the orientational trap, the torsional modulus can be determined.
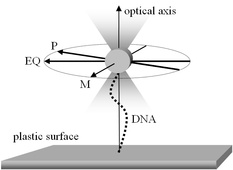
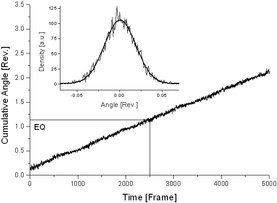
Optical trapping arrangement (left). Notations: P: polarization plane of the trapping light, M: direction corresponding to the zero torsional strain of the molecule, EQ: equilibrium orientation of the disc. Typical raw data showing the orientation of the disc during data collection (right). The plane of polarization is slowly rotated during the measurements in order to minimize the effects of optical anisotropic errors. The average of cumulative angles defines the equilibrium orientatation EQ corresponding to the middle of the period. The calibration of the optical trap is based on the analysis of rotational Brownian fluctuations of the disc (insert). Results In their statistical mechanics model of twist-storing polymers, Moroz and Nelson showed that the effective torsional modulus (and so the Ceff effective twist persistence length) of the molecule depends on its (fixed) relative extension. The less the extension (pulling force) of the chain, the stronger the thermal bend fluctuations resulting in a lower measurable torsional stiffness. The effect has been calculated quantitatively and shows a dependence on the ratio of the twist and bend persistence lengths C/A and the temperature. The smaller the bend persistence length A compared to the twist persistence length C, the more reduced the measurable (effective) persistence length of the molecule. At the maximum relative extension (when no bending is allowed) Ceff is equal to C and characterizes the microscopic (local) twist stiffness of the molecule. In our experiments the effective torsional modulus of DNA has been measured at 0.5 and 0.75 relative extensions. When fitting the experimental values to the Moroz-Nelson model we obtain a value of 420 ± 43 pN nm2 for the local torsional modulus of dsDNA (equivalent to a twist persistence length C of 102 ± 10 nm, using a value of 50 nm for the bend persistence length A). The value for the torsional modulus basically identical to that obtained recently by the Bustamante group (440 ± 40 pN nm2 and 410 ± 30 pN nm2 measured at 15 pN and 45 pN tensions, respectively). However these values are significantly higher than those reported earlier from supercoiled single-molecule DNA stretching experiments using magnetic tweezers. We note that these experiments were also evaluated by Moroz and Nelson yielding a value of 109 nm for the local twist persistence length in a remarkable agreement with recent experimental data (the Bustamante group and us). When using the treatment of Moroz and Nelson for the magnetic tweezers experiments and our work, all available results are consistent, providing strong additional support for this model. Reference: László Oroszi, Péter Galajda, Huba Kirei, Sándor Bottka, and Pál Ormos, Direct Measurement of Torque in an Optical Trap and Its Application to Double-Strand DNA, Phys. Rev. Lett. 97, 058301 (2006). |