|
|
|
|
|
|
 | |
Absorption kinetics on proteins of bioenergetics |
Principal Investigator: László Zimányi
Bacterial retinal proteins function as either signal transducers (e.g. sensory rhodopsin) or ion pumps (e.g.
bacteriorhodopsin and halorhodopsin). These 7 transmembrane a-helical proteins bind a retinal chromophore covalently.
In the ion pumps light energy absorbed by the retinal is converted into transmembrane electrochemical gradient by a
sequence of events comprising of retinal isomerization, proton (in the case of bacteriorhodopsin) transfer between
various sites within the protein and between the protein and the aqueous medium, and gradually spreading protein
conformational change. The molecular steps of this cyclic reaction sequence are followed by time resolved kinetic
absorption spectroscopy.
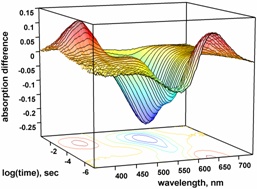
Chemometric methods, such as singular value decomposition with exponential fit assisted self-modeling (SVD-EFASM), are
applied to obtain the most likely spectra of the photocycle intermediates and their kinetics. Various reaction schemes
are fitted to the kinetic data.
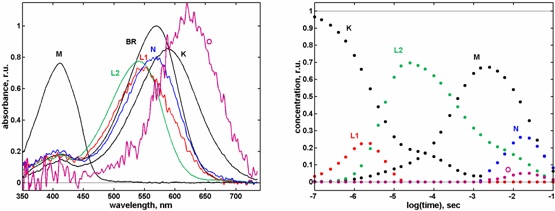
Our improved methods have led to the discovery of consecutive, spectrally different L intermediates in the wild type
bacteriorhodopsin photocycle.
References:
Zimányi, L., Kulcsár, Á., Lanyi, J.K., Sears, D.F. and Saltiel, J. 1999. Singular value decomposition with self-modeling applied to determine bacteriorhodopsin intermediate spectra: Analysis of simulated data. Proc. Natl. Acad. Sci. USA 96:4408-4413
Zimányi, L., Kulcsár, Á., Lanyi, J.K., Sears, D.F. and Saltiel, J. 1999. Intermediate spectra and photocycle kinetics of the Asp96 ? Asn mutant bacteriorhodopsin determined by singular value decomposition with self-modeling. Proc. Natl. Acad. Sci. USA 96:4414-4419.
Kulcsár, Á., Saltiel, J. and Zimányi, L. 2001. Dissecting the photocycle of the bacteriorhodopsin E204Q mutant from kinetic multichannel difference spectra. Extension of the method of singular value decomposition with self-modeling to five components. J. Am. Chem. Soc. 123:3332-3340
Zimányi, L. 2002. Kinetic multichannel spectroscopy of biological molecules: Decomposition of the spectral matrix. Biopolymers (Biospectroscopy) 67:263-266
Zimányi, L. 2004. Analysis of the bacteriorhodopsin photocycle by singular value decomposition with self-modeling: a critical evaluation using realistic simulated data. J. Phys. Chem. B 108: 4199-4209
Zimányi, L., Saltiel, J., Brown, L.S. and Lanyi, J.K. 2006. A Priori Resolution of the Intermediate Spectra in the Bacteriorhodopsin Photocycle: The Time Evolution of the L Spectrum Revealed. J. Phys. Chem. A 110(7):2318-2321
| |


